|
Oligo Constructs Research
Basic Overview
In this research, Dr. Jennifer Weller and myself were replicating and extending the reseach of one of her graduate students, Saeed Khoshnevis.
The goal was to study the effects of structures in DNA on the accuracy that DNA sequencers achieve. We did this using small pieces of DNA that
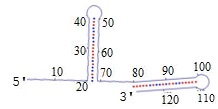
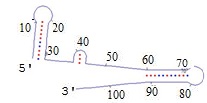
have a known sequence and structure, like those shown below.
Since A bonds to T and C bonds to G, a piece of DNA can loop back on itself - this is called a hairpin.
We made a very small amount of the DNA we wanted (our Target) using short pieces of DNA that we had chemically synthesized. To make more of the Target we used PCR to make copies and then
purified the products we wanted. Next we sequenced the DNA fragments in an Ion Torrent PGM sequencer and compared the order of A,C,T and G (the sequence results) to what we knew we had purified.
We noticed that some of the DNA sequence near the hairpin structure appeared to be missing, which means that the Targets weren't sequenced properly. This allowed us to determine that structure
does in fact affect sequencing results, as we hypothesized.
|
Links To More Detailed Descriptions
Should you be wondering why I started this reseach in the first place, and you are well versed in genomics terms (aka Science Jargon,and I'm not kidding, you really have to understand this vocabulary
to know what this publication is talking about), then this is where our research started. This journal artible discusses differences that were found in the sequence of individual who was part of a Korean "Whole-Genome Sequence"
study. His DNA was found to be "missing" a surprisingly large number of short pieces of DNA throughout his genome; our question was whether there a structure might be interfering with the sequencing process. (A Highly Annotated Whole-Genome Sequence of a Korean Individual).
Briefly, in 2009 a group of researchers wanted to see if they could pinpoint an individuals origins based on his genomic DNA. So they created and sequenced a genome library from a Korean indiviual, designated AK1,
and compared his sequence to a reference. Since AK1 was missing some potentially important coding DNA compared to the reference, the researchers tried to see if the deletions were associated with different genetc illnesses, but failed to find any.
However, our research shows that if there are hairpins in the DNA in these regions sequencing methods might misread the DNA and we will interpret it incorrectly.
Mistakes like this could mislead researchers into investing a lot of time and money pursuing non-existent problems.
Basic Information: If you would like an explanation of the basic processes I used, click here.
Technical Information: I am currently working on a page that includes some of the sequence data and how we analyzed it.
|
Walsh Project Main Page
|
|